Randomized SMILES Strings Improve the Quality of Molecular Generative Models
The paper Randomized SMILES Strings Improve the Quality of Molecular Generative Models has been published as a preprint on ChemRxiv.
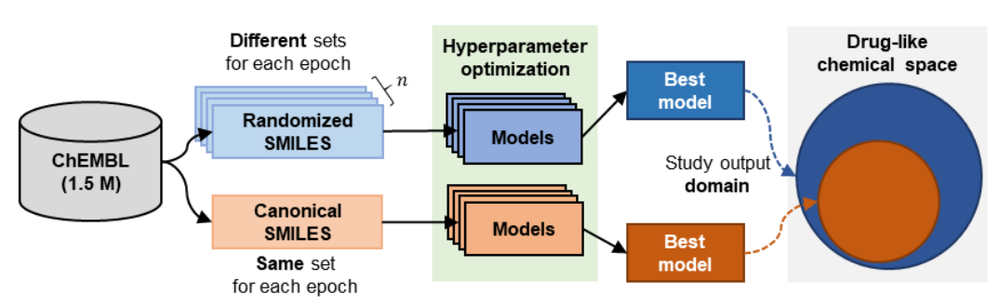
Recurrent Neural Networks (RNNs) trained with a set of molecules represented as unique (canonical) SMILES strings, have shown the capacity to create large chemical spaces of valid and meaningful structures. Herein we perform an extensive benchmark on models trained with subsets of GDB-13 of different sizes (1 million , 10,000 and 1,000), with different SMILES variants (canonical, randomized and DeepSMILES), with two different recurrent cell types (LSTM and GRU) and with different hyperparameter combinations. To guide the benchmarks new metrics were developed that define the generated chemical space with respect to its uniformity, closedness and completeness. Results show that models that use LSTM cells trained with 1 million randomized SMILES, a non-unique molecular string representation, are able to generate larger chemical spaces than the other approaches and they represent more accurately the target chemical space. Specifically, a model was trained with randomized SMILES that was able to generate almost all molecules from GDB-13 with a quasi-uniform probability. Models trained with smaller samples show an even bigger improvement when trained with randomized SMILES models. Additionally, models were trained on molecules obtained from ChEMBL and illustrate again that training with randomized SMILES lead to models having a better representation of the drug-like chemical space. Namely, the model trained with randomized SMILES was able to generate at least double the amount of unique molecules with the same distribution of properties comparing to one trained with canonical SMILES.
Author(s): Josep Arús-Pous, Simon Johansson, Oleksii Ptykhodko, Esben Jannik Bjerrum, Christian Tyrchan, Jean-Louis Reymond, Hongming Chen, Ola Engkvist