Exploring the GDB-13 chemical space using deep generative models
The paper Exploring the GDB-13 chemical space using deep generative models has been published by the Journal of Cheminformatics.
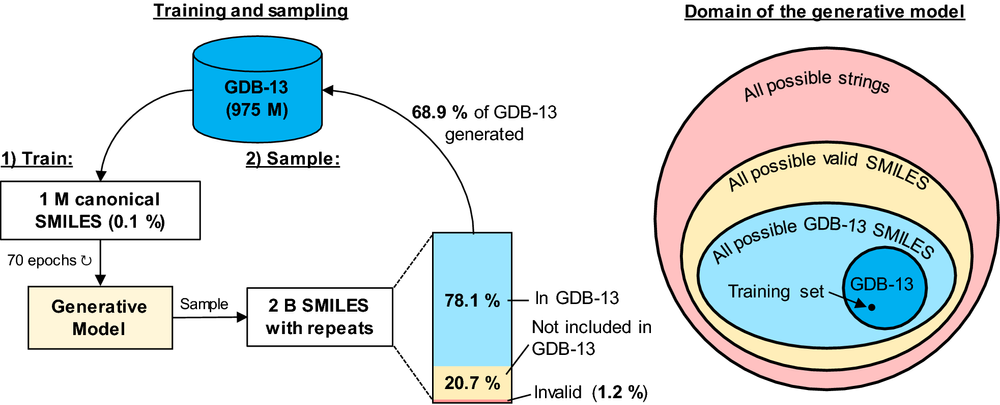
Recent applications of recurrent neural networks (RNN) enable training models that sample the chemical space. In this study we train RNN with molecular string representations (SMILES) with a subset of the enumerated database GDB-13 (975 million molecules). We show that a model trained with 1 million structures (0.1% of the database) reproduces 68.9% of the entire database after training, when sampling 2 billion molecules. We also developed a method to assess the quality of the training process using negative log-likelihood plots. Furthermore, we use a mathematical model based on the “coupon collector problem” that compares the trained model to an upper bound and thus we are able to quantify how much it has learned. We also suggest that this method can be used as a tool to benchmark the learning capabilities of any molecular generative model architecture. Additionally, an analysis of the generated chemical space was performed, which shows that, mostly due to the syntax of SMILES, complex molecules with many rings and heteroatoms are more difficult to sample.
Author(s): Josep Arús-Pous, Thomas Blaschke, Silas Ulander, Jean-Louis Reymond, Hongming Chen, Ola Engkvist