A View on Molecular Complexity from the GDB Chemical Space
Check out our latest publication A View on Molecular Complexity from the GDB Chemical Space in JCIM!
Abstract
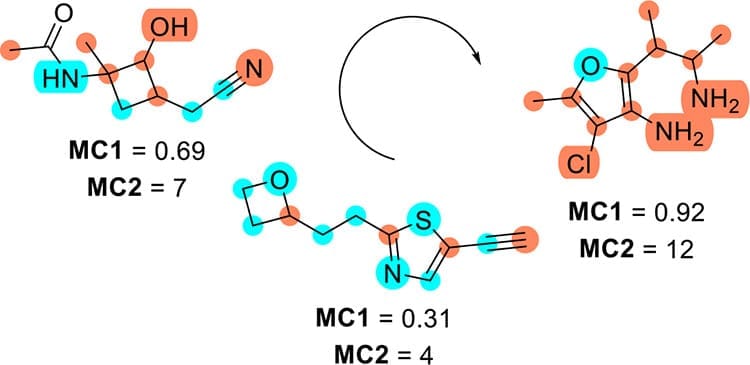
One recurring question when choosing which molecules to selectfor investigation is that of molecular complexity: is there a price to pay forcomplexity in terms of synthesis difficulty, and does complexity have anything todo with biological properties? In the chemical space of small organic moleculesenumerated from mathematical graphs in the GDBs (Generated DataBases), mostcompounds are too complex and challenging for synthesis despite containing onlystandard functional groups and ring types. For these GDB molecules, we find thatan increasing fraction (MC1) or number (MC2) of non-divalent nodes in themolecular graph represent simple measures of molecular complexity, which weinterpret in terms of potential synthesis difficulties. We also show that MC1 and MC2 are applicable to commercial screeningcompounds (ZINC), bioactive molecules (ChEMBL) and natural products (COCONUT) and compare them with previouslyreported measures of molecular complexity and synthetic accessibility.
Author(s) Ye Buehler and Jean-Louis Reymond