Molecular Framework Analysis of the Generated Database GDB-13s
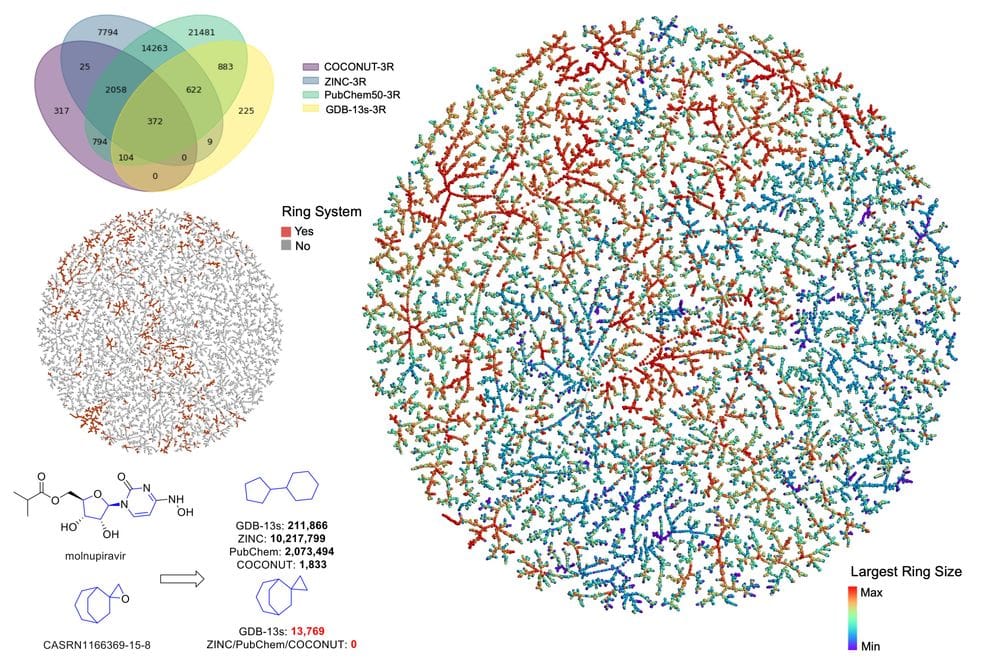
The paper on Bemis and Murcko molecular frameworks of GDB-13s molecules is now published in JCIM! Here we assessed the originality of GDB and found out that many exclusive molecular frameworks in GDB might be the most relevant targets for synthetic chemistry aiming at innovative molecules.
Check it out: Molecular Framework Analysis of the Generated Database GDB-13s
Abstract:
The generated databases (GDBs) list billions of possible molecules from systematic enumeration following simple rules of chemical stability and synthetic feasibility. To assess the originality of GDB molecules, we compared their Bemis and Murcko molecular frameworks (MFs) with those in public databases. MFs result from molecules by converting all atoms to carbons, all bonds to single bonds, and removing terminal atoms iteratively until none remain. We compared GDB-13s (99,394,177 molecules up to 13 atoms containing simplified functional groups, 22,130 MFs) with ZINC (885,905,524 screening compounds, 1,016,597 MFs), PubChem50 (100,852,694 molecules up to 50 atoms, 1,530,189 MFs), and COCONUT (401,624 natural products, 42,734 MFs). While MFs in public databases mostly contained linker bonds and six-membered rings, GDB-13s MFs had diverse ring sizes and ring systems without linker bonds. Most GDB-13s MFs were exclusive to this database, and many were relatively simple, representing attractive targets for synthetic chemistry aiming at innovative molecules.
Author(s): Ye Buehler and Jean-Louis Reymond